Simulating an Ising Model#
In this page, you will write Monte Carlo code to sample configurations from the Ising model with the correct probability.
Minimal background#
The Ising model#
The Ising model is one of the drosophila of physics. At first look, it’s a simple model of crystalline materials which attributes magnetism to the orientation of magnetic moments on a lattice, summarized by the Hamiltonian or energy
The spins have two orientations, \(s_{i} = \pm 1\), which only interact with their *nearest neighbors. There are more complicated versions of the Ising model (which we won’t use in the main part of the assignment) whose energy terms have a different value \(J_{ij}\) for every bond \((i,j)\) - i.e. \(J_{ij} s_i s_j\) or an additional term for a magnetic field \(\sum_i h_i s_i\).
A configuration of an Ising model is a specification of every moment’s orientation — an assignment of values to the \(s_i\) variable for all moments in the system. For our purposes, configurations are ‘snapshots’ of the system which our simulator will hold in memory, like a vector containing \(\pm 1\) in its \(i\)th entry to specify whether moment \(i\) points up or down.
Statistical Mechanics#
There are a couple facts we need to know about statistical mechanics.
(1) We can specify configurations of our system \(c.\) In the case of an Ising model, the configuration is the orientation of each spin.
(2) Each configuration \(c\) has an energy \(E(c)\). We’ve already seen this for the Ising model.
(3) For classical systems in contact with an energy reservoir of temperature \(T\), the probability \(P\) of finding the system in configuration \(c\) is
\(P(c) = \frac{e^{-\beta E(c)}}{\sum_c e^{-\beta E(c)}}\)
where \(E(c)\) is the configuration’s energy and \(\beta \equiv 1/(kT)\) with \(k\) Boltzmann’s constant and \(T\) is the temperature. We will work in units where \(k=1.\)
Markov Chains#
We have just learned that statistical mechanics tells us that if we look at the system, each configuration \(c\) appears with probability \(P(c)\sim \exp(-\beta E(c))\). Our first goal is to implement an algorithm which gives us configurations \(c\) with probability \(P(c)\).
One such algorithm is markov chain Monte Carlo.
A Markov chain is a process where you are at some state \(c\) (i.e. a configuration) and then you choose another state \(c'\) with a probability that only depends on \(c\). In other words, you can’t use the fact that previously you were at configuration \(b\) to decide the probability \(P(c \rightarrow c')\) you’re going to go from \(c\) to \(c'\). This process is called a memoryless process.
As long as you do a (non-pathalogical) random walk in a memoryless way, you’re guaranteed that, after walking around enough, that the probability you are in configuration \(c\) is some \(\pi(c)\). \(\pi\) is called the stationary distribution. Different markov chains will have different stationary distributions. A Markov chain is non-pathological if:
It is aperiodic; it doesn’t cycle between configurations in a subset of the system’s full configuration space.
It is connected; given two configurations, there is a path with non-zero probability that the chain could (but not necessarily will) follow to get from one to the other.
To simulate the Ising model, we wish to build a markov chain which has the stationary distribution \(\pi(c) \sim \exp(-\beta E_c)\). We will do so with a very famous algorithm which lets us build a Markov chain for any \(\pi(c)\) we want.
The Metropolis-Hastings algorithm#
If you know your desired stationary distribution, the Metropolis-Hastings algorithm provides a canonical way to generate a Markov chain that produces it. The fact that you can do this is amazing! The steps of the algorithm are:
Start with some configuration \(c\)
Propose a move to a new trial configuration \(c'\) with a transition probability \(T(c\rightarrow c')\)
Accept the move to \(c'\) with probability \(\min \left(1, \frac{\pi(c')}{\pi(c)}\frac{T(c'\rightarrow c)}{T(c \rightarrow c')} \right)\)
The first step is straightforward, but the latter two deserve some explanation.
How \(T(c\rightarrow c')\) is picked: Choosing a procedure for movement between configurations is the art of MCMC, as there are many options with different strengths and weaknesses. In the simplest implementations of the Metropolis algorithm we choose a movement procedure where forward and reverse moves are equiprobable, \(T(c \rightarrow c') = T(c' \rightarrow c)\).
Question to think about: How do equiprobable forward and reverse moves simplify the Metropolis algorithm?
Acceptance condition: To construct a Markov chain which asymptotes to the desired distribution \(\pi\), the algorithm must somehow incorporate \(\pi\) in its walk through configuration space. Metropolis does this through the acceptance ratio
Notice how the first factor in \(\alpha\) is the relative probability of the configurations \(c'\) and \(c\). Assuming that \(T(c \rightarrow c')=T(c' \rightarrow c)\), we have that \(\alpha > 1\) indicates that \(c'\) is more likely than \(c\), and the proposed move is automatically accepted. When \(\alpha < 1\), \(c'\) is less probable than \(c\), and the proposed move is accepted with probability \(\alpha\).
Question to think about: Why are moves with \(\alpha < 1\) accepted with probability \(\alpha\), rather than outright rejected?
States and parameters#
For our simulation, we are going to work on a two-dimensional lattice. This leaves us with an energy
where \((a,b) \in \{(1,0) , (0,1), (-1,0), (0,-1)\}\)
For this project we will be working on a \(L \times L\) grid (where \(L\) is 3 or 27 or 81).
We are going to represent the state of our spins by a \(L \times L\) array which can have either a -1 (spin-down) or 1 (spin-up). To set up a random initial conidition you can do np.random.choice([1,-1], (L,L)) in python. In C++, there are two approaches to storing the two dimensional array. One approach is to use the STL: vector<vector<int> > spins. Another option is to use Eigen (see here) where you will use MatrixXi.
This approach will work for square grids (and is what you want to do for this project!). In some cases, you want a more complicated lattice (triangular, pyrochlore) and have different parameters for \(J_{ij}\) and \(h_i\). In that case, instead of storing it as a grid you can alternatively store it as a graph. This below is just for your edification for future uses.
Storing your configuration as a graph.
A more general way to store your configuration is to put your spins (and magnetic field) on nodes of a graph and the coupling constants \(J_{ij}\) as edges.
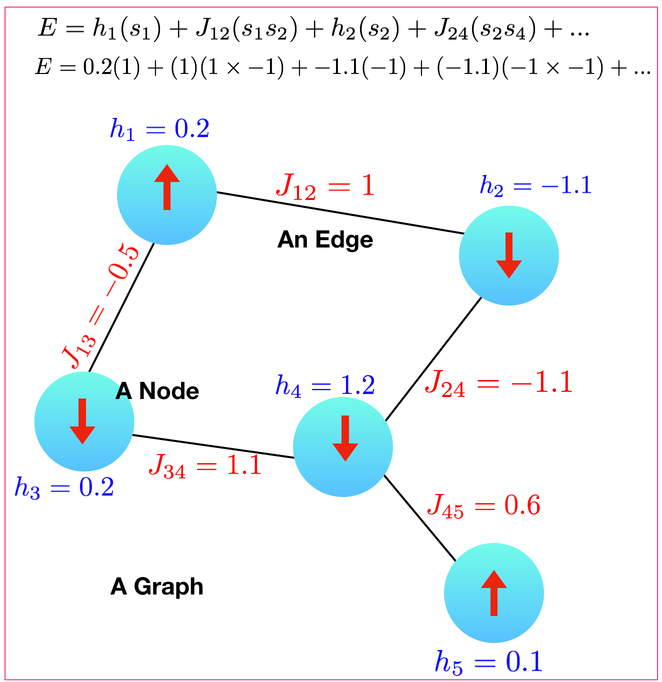
A graph has a bunch of nodes (each node \(i\) has a state \(s_i\) which is either spin-up or spin-down and has a magnetic field \(h_i\)) and a bunch of edges (each edge has a \(J_{ij}\) on it). Then, if you want a grid you can just choose a graph that is grid-like.
Now there are two questions: how do you represent this inside your code and how do you get the code to know about the graph that you want.
Representing your spins: Here is what I did in my code. I wrote an Ising class which has two vectors which represent the state of the nodes and their magnetic fields (i.e. vector<int> spins and vector<double> h). Then, I stored a neighbor vector. The i’th element in the neighbor vector stores a list (or vector) of neighbors with their respective coupling constants.
In other words, for the graph above, you want to be storing the following inside your computer:
Node: (neighbor, J) (neighbor, J) (neighbor, J) ...
1: (2, 1) (3,-0.5)
2: (1,1) (4,-1.1)
3: (1,-0.5) (4,1.1)
4: (2,-1.1) (3,1.1) (5,0.6)
5: (4,0.6)
I stored this as vector<vector<pair<int, double> > > neighbors where the pair<int,double> stores the node you are a neighbor to and the weight.
Setting up your graph: Setting up your Ising model this way gives you a lot of flexibility. But now you need some way to tell your code what particular graph you want (maybe the one above or a grid, etc).
I recommend doing this by reading two files. A “spins.txt” which tells you the value of \(h_i\) for spin \(i\) and a “bonds.txt” which tells you which spins are connected and what their coupling constants are.
For the first graph above: bonds.txt
1 2 1.0
1 3 -0.5
3 4 1.1
2 4 -1.1
4 5 0.6
spin.txt
1 0.2
2 -1.1
3 0.2
4 1.2
5 0.1
Then just write some sort of Init() function for your Ising model to read in the graph. Also, go ahead and write yourself a python code that generates the relevant graph for a two-dimensional Ising model on a square grid. Now, any time you want to run a different Ising model, you just have to change these files.
Computing the Energy#
The next step in putting together your Ising model is learning how to compute the energy \(E(c)\) of a configuration. Write some code (i.e def Energy(spins)) which allows you to do this.
In addition to computing the energy for a single configuration, it’s going to be critical to be able to compute the energy difference between two configurations that differ by a single spin flip (i.e. def deltaE(spin_to_flip))
Why is this? Because the acceptance criterion we are going to use is the ratio of \(\exp[-\beta E_\textrm{new}]/\exp[-\beta E_\textrm{old}]=\exp[-\beta \Delta E]\).
Go ahead and make this function. Make sure that the function takes time \(O(1)\) and not time \(O(N)\) (i.e. don’t call Energy(spins) twice).
Comment about computational complexity: One of the themes of this class is to think about the scaling of your algorithms (like we did thinking about various options for quantum computing). Let’s do this for computing an energy difference. One thing you can do is compute the energy, flip the spin, compute the energy, and subtract. How long does this take? It takes \(O(N + B)\) time where \(N\) is the number of spins and \(B\) is the number of bonds. Find a way to do it with a time that is proportional to \(O(1)\).
At this point you should be able to store a configuration of spins and compute energy and energy differences on them.
Testing
Set up a \(3 \times 3\) grid and compute the energy of some spin configuration both by hand and with your code to make sure it’s the same
Compute the energy difference of flipping a single spin both by
using your
Energy()function before and after a spin flip.using your
deltaE()function.
Both approaches for computing the energy should be the same. Another reasonable test is to compute the energy of a number of configurations that you can evaluate by hand (all spin-up, etc) and verify that it works.
Writing MCMC#
We now want to put together the Monte Carlo. The basic operation, which we call a step is to do the following (many times):
Choose a new configuration \(c'\) with some probability \(p(c \rightarrow c')\) given the current configuration \(c\). The simplest method is to choose a random spin and flip it. In this case both \(T(c \rightarrow c') = T(c' \rightarrow c) = 1/N\) (where \(N\) is the number of spins). Therefore the ratio of \(T\) cancel in the acceptance probability \(\alpha\)).
Evaluate the difference in energy \(\Delta(c,c) \equiv E(c') - E(c)\)
If \(\frac{T(c' \rightarrow c)}{T(c \rightarrow c')} \exp[-\beta \Delta(c,c')]>\textrm{ran}(0,1)\) then accept the new configuration \(c'\). Otherwise leave the old configuration \(c\)
We will call a sweep doing \(N\) steps.
You will typically need to do a certain number (maybe 20) sweeps before you should start collecting data. This is called the equilibration time and is required because you need the Markov Chain to forget about your initial condition.
Then you can take a measurement every sweep. There is no need to measure more often then this (and doing so will just slow down the simulation).
This is going to look something like this (in c++)
for (int sweep=0;sweep<10000;sweep++){
for (int i=0;i<N;i++){
//Pick a random spin and try to flip it
}
//measure your histogram or energy, etc.
}
or (in python)
for sweep in range(0,10000):
for step in range(0,N):
#flip spins
# measure
Grading
For your \(3 \times 3\) grid, run your Monte Carlo and make a histogram of the configurations. To do this, you should take a snapshot of your spin-configuration every sweep. Your spin-configuration can be flattened down to nine spins (i.e. \(\uparrow, \downarrow, \uparrow ... \uparrow)\). This can be turned into a nine-bit binary number (\(101..1\)) which can then be turned into an integer from 0 to \(2^9-1.\) You then have an integer for every sweep from which to make a histogram.
Then, compare this histogram against the theoretical values - i.e you can compute the energy of all \(2^9\) configurations (using python) and then you can use the formula \(\exp[-\beta E(c)]/Z\) (where \(Z\) is the normalization) to make the theory graph from this.
You should have both graphs on top of each other on the same plot (maybe one with points and one with lines) to see that they agree. There is going to be some error bar on your Monte Carlo data so they don’t have to match exactly.
Note, that this is a great way to test your code. It’s only going to work with a small number (i.e. 9) of spins. If you tried to do this with 25 spins, you would need much too large a histogram.
How often should you take snapshots (or more generically compute observables)
The promise of MCMC is that after a “long enough time” \(T\), you should get a random sample (we unfortunately don’t know \(T\) a-priori - it’s at least one sweep but could be many sweeps). Roughly you want to measure an observable or add to your histogram every \(T\) sweeps.
Q: What happens if you take snapshots too infrequently? You end up just wasting some time. The samples are still independent.
Q: What happens if you take snapshots too frequently? That’s a little more subtle. If you take snapshots too soon then you are in trouble. This means that you should wait a fair amount of time before you start measuring snapshots.
If you wait enough time for the first snapshot and then take measurements more frequently then you are still ok. It’s just you have to be careful with your errorbars because subsequent snapshots may be correlated.
Later on we will see how to estimate \(T\). For this simulation,let’s just assume that \(T\) is about a sweep. Wait 10 sweeps before you begin measuring snapshots. Then take a snapshot every sweep. Error bars for your histogram: If you have \(k\) samples in a bin then your error should be \(\sqrt{k}\).
If you are using C++, your code will be very efficient (although make sure you are compiling with -O3 or it will be much too slow!)
In python unfortunately, this will turn out to be very slow especially if you are using for loops.
The code will be correct but it will take a very long time (this doesn’t matter and won’t necessarily show up for the \(3 \times 3\) system but will mean you have to wait a long time for the larger systems you will do later in this assignment).
Speeding Up Your Code#
In order for you to be able to finish your assignment, your code will need to be sufficiently fast. If you are working in C++ or Julia, it will be natively fast enough. If you are using python, the simplest way to make it fast (and the way you should use) is to just-in-time compile the functions that are slow. This can be done using numba. See
numba for instructions. Numba can be slightly tempremental so you may have to modify your code in some ways so that it can compile through the functions.
Before numba was working well enough, we had a slower approach which used numpy to avoid for-loops. You can see this older approach on this page (but you shouldn’t do this if you don’t have a good reason).
In terms of speed, roughly what you should find for 10,000 sweeps for an \(81 \times 81\) lattice:
Naive Python: ~13min 18s
Python with numpy avoiding for loops: ~56.3 s
Python with numba on top of the naive approach: ~13.5 s
Once you’ve implemented this with numba, each step should be very fast. That will be sufficient to complete the assignment. There is one thing that is still somewhat inefficient. As you approach the critical temperature, the autocorrelation time goes up. This causes you to need to do a lot of sweeps before you get an independent sample. If you want to overcome this, you can implement a cluster move (Extra Credit: 5+5 points)
Coupling From the Past#
Extension and Extra Credit (10 points)
By writing a Markov Chain Monte Carlo, you’ve produced a code which samples from the Boltzmann distribution for an Ising model - at least if we look away for long enough. Unfortunately, we don’t know how long that necessarily needs to be. In practice, this is rarely a problem. Nonetheless, it would be satisfying if there was an algorithm that exactly sampled from the Boltzmann distribution of the Ising model with no heurestics or additional approximations. There is such an algorithm: coupling from the past. This algorithm is on my list of 10 algorithms I didn’t believe could accomplish what it was claiming when I first heard it (automatic differentiation is also on that list).